GSoC 2016: Port NMatrix to JRuby
I have been working on “Port NMatrix to JRuby” as my GSoC project. I am pleased to announce that JRuby is ready for Nmatrix users.
NMatrix, a linear algebra library wraps Apache Commons Maths for its core functionalities. By the end of GSoC, I have been able to implement NMatrix for dense matrices with double and object ( ruby objects ) data type. I have also worked on porting mixed-models gem to JRuby which heavily uses NMatrix at its core.
This blog post summarizes my work on the project with Sciruby, and reports the final status of the project.
Proposal
The proposal application can be found here.
Code Commits
https://github.com/prasunanand/nmatrix/commits/jruby_port
Storing n-dimensional matrices as flat arrays
The major components of a NMatrix is its shape, elements, dtype and stype. Any nmatrix when initialised, the elements are stored in flat arrays. ArrayRealVector class is used to store the elements.
@s stores the elements, @shape stores the shape of array, while @dtype and @stype store the data type and storage type respectively. currently, we have nmatrix-jruby implemented for only double matrices.
NMatrix-MRI uses @s which is an object containing elements, stride, offset as in C, we need to deal with the memory allocation for the arrays.

Slicing and Rank
Implementing slicing was the toughest part of NMatrix-JRuby implementation. NMatrix@s stores the elements of a matrix as a flat_array. The elements along any dimension are accessed with the help of the stride. NMatrix#get_stride calculates the stride with the help of the dimension and shape and returns an array.
def get_stride(nmatrix)
stride = Array.new()
(0...nmatrix.dim).each do |i|
stride[i] = 1;
(i+1...dim).each do |j|
stride[i] *= nmatrix.shape[j]
end
end
stride
end
NMatrix#[] and NMatrix#[]= are thus able to read and write the elements of a matrix. NMatrix#MRI uses the @s object which stores the stride when the nmatrix is initialised.
NMatrix#[] calls the #xslice operator which calls #get_slice operator that use the stride to find out whether we are accessing a single element or multiple elements. If there are multiple elements #dense_storage_get then returns an NMatrix object with the elements along the dimension.
NMatrix-MRI differs from NMatrix-JRuby implementation as it makes sure that memory is properly utilized as the memory needs to be properly garbage collected.
def xslice(args)
result = nil
if self.dim < args.length
raise(ArgumentError,"wrong number of arguments\
(#{args} for #{effective_dim(self)})")
else
result = Array.new()
slice = get_slice(@dim, args, @shape)
stride = get_stride(self)
if slice[:single]
if (@dtype == :object)
result = @s[dense_storage_get(slice,stride)]
else
s = @s.toArray().to_a
result = @s.getEntry(dense_storage_get(slice,stride))
end
else
result = dense_storage_get(slice,stride)
end
end
return result
end
NMatrix#[]= calls the #dense_storage_set operator which calls #get_slice operator that use the stride to find out whether we are accessing a single element or multiple elements. If there are multiple elements #set_slice recursively sets the elements of the matrix then returns an NMatrix object with the elements along the dimension.
def dense_storage_set(slice, right)
stride = get_stride(self)
v_size = 1
if right.is_a?(NMatrix)
right = right.s.toArray.to_a
end
if(right.is_a?(Array))
v_size = right.length
v = right
if (dtype == :object)
# nm_register_values(reinterpret_cast<VALUE*>(v), v_size)
end
(0...v_size).each do |m|
v[m] = right[m]
end
else
v = [right]
if (@dtype == :object)
# nm_register_values(reinterpret_cast<VALUE*>(v), v_size)
end
end
if(slice[:single])
# reinterpret_cast<D*>(s->elements)[nm_dense_storage_pos(s, slice->coords)] = v;
pos = dense_storage_pos(slice[:coords],stride)
if @dtype == :object
@s[pos] = v[0]
else
@s.setEntry(pos, v[0])
end
else
v_offset = 0
dest = {}
dest[:stride] = get_stride(self)
dest[:shape] = shape
# dest[:elements] = @s.toArray().to_a
dense_pos = dense_storage_pos(slice[:coords],stride)
slice_set(dest, slice[:lengths], dense_pos, 0, v, v_size, v_offset)
end
end
Enumerators
NMatrix-MRI uses the C code for enumerating the elements of a matrix. However, the NMatrix-JRuby uses pure Ruby code. Currently, all the enumerators for dense matrices with real data-type have been implemented and they are properly functional. We haven’t implemented enumerators for objects currently.
def each_with_indices
nmatrix = create_dummy_nmatrix
stride = get_stride(self)
offset = 0
#Create indices and initialize them to zero
coords = Array.new(dim){ 0 }
shape_copy = Array.new(dim)
(0...size).each do |k|
dense_storage_coords(nmatrix, k, coords, stride, offset)
slice_index = dense_storage_pos(coords,stride)
ary = Array.new
if (@dtype == :object)
ary << self.s[slice_index]
else
ary << self.s.toArray.to_a[slice_index]
end
(0...dim).each do |p|
ary << coords[p]
end
# yield the array which now consists of the value and the indices
yield(ary)
end if block_given?
nmatrix.s = @s
return nmatrix
end
Two Dimensional Matrices
Linear algebra is mostly about two-dimensional matrices. In NMatrix, when performing calculations in a two-dimensional matrix, a flat array is converted to a two-dimensional matrix. A two-dimensional matrix is stored as a BlockRealMatix or Array2DRowRealMatrix. Each of them has its own advantages.
Getting a two-d-matrix

public class MatrixGenerator
{
public static double[][] getMatrixDouble(double[] array, int row, int col)
{
double[][] matrix = new double[row][col];
for (int index=0, i=0; i < row ; i++){
for (int j=0; j < col; j++){
matrix[i][j]= array[index];
index++;
}
}
return matrix;
}
}
Flat a two-d matrix
public class ArrayGenerator
{
public static double[] getArrayDouble(double[][] matrix, int row, int col)
{
double[] array = new double[row * col];
for (int index=0, i=0; i < row ; i++){
for (int j=0; j < col; j++){
array[index] = matrix[i][j];
index++;
}
}
return array;
}
}
Why use java method instead of Ruby method?
-
Memory Usage and Garbage Collection => A scientific library is memory intensive and hence, every step counts. JRuby interpreter doesn’t need to dynamically guess the data type and uses less memory, i.e around 10 times. If the memory is properly utilized; when the GC kicks in, it has to clear less memory and improves the speed.
-
Speed => Using java method greatly improves the speed around 1000 times, when compared to using ruby method.
Operators
All the operators from NMatrix-MRI have been implemented except moduli. The binary operators were easily implemented through Commons Math Api.
def +(other)
result = create_dummy_nmatrix
if (other.is_a?(NMatrix))
#check dimension
raise(ShapeError, "Cannot add matrices with different dimension")\
if (@dim != other.dim)
#check shape
(0...dim).each do |i|
raise(ShapeError, "Cannot add matrices with different shapes") \
if (@shape[i] != other.shape[i])
end
result.s = @s.copy.add(other.s)
else
result.s = @s.copy.mapAddToSelf(other)
end
result
end
Trigonometric, exponentiation and log operators with a singular argument i.e. matrix elements were implemented using mapToSelf method that that takes univariate function as an argument. mapToSelf maps every element of ArrayRealVector to the Univate operator passed to it and returns self object.
def sin
result = create_dummy_nmatrix
result.s = @s.copy.mapToSelf(Sin.new())
result
end
NMatrix#method(arg) was implemented using Bivariate functions provided by Commons-Maths and Java Maths library.
def gamma
result = create_dummy_nmatrix
result.s = ArrayRealVector.new MathHelper.gamma(@s.toArray)
result
end
import org.apache.commons.math3.special.Gamma;
public class MathHelper{
...
public static double[] gamma(double[] arr){
double[] result = new double[arr.length];
for(int i = 0; i< arr.length; i++){
result[i] = Gamma.gamma(arr[i]);
}
return result;
}
...
}
Decomposition
NMatrix-MRI relies on LAPACKE and ATLAS for matrix decomposition and solve functionalities. Apache Commons Math provides a different set of API for decomposing a matrix and solving an equation. for-example potrf and other LAPACKE specific functions have not been implemented as they are not required at all.
Calculating determinant in NMatrix is tricky where a matrix is reduced either a Lower or Upper matrix and the diagonal elements of the matrix are multiplied to get the result. Also, the correct sign of the result whether positive or negative is taken into account, while calculating the determinanat. However, NMatrix-JRuby uses commons-math api to calculate the determinant.
def det_exact
if (@dim != 2 || @shape[0] != @shape[1])
raise(ShapeError, "matrices must be square to have a determinant defined")
return nil
end
to_return = LUDecomposition.new(self.twoDMat).getDeterminant()
end
Given below is the code, that shows how Cholesky decomposition has been implemented by using Commons Math API. Similarly, LU Decomposition and QR factorization have been implemented.
Cholesky Decomposition
def factorize_cholesky
cholesky = CholeskyDecomposition.new(self.twoDMat)
l = create_dummy_nmatrix
twoDMat = cholesky.getL
l.s = ArrayRealVector.new(ArrayGenerator.getArrayDouble\
(twoDMat.getData, @shape[0], @shape[1]))
u = create_dummy_nmatrix
twoDMat = cholesky.getLT
u.s = ArrayRealVector.new(ArrayGenerator.getArrayDouble\
(twoDMat.getData, @shape[0], @shape[1]))
return [u,l]
end
Cholesky Decomposition for an NMatrix-JRuby requires the matrix to be square matrix.
LUDecomposition
def factorize_lu with_permutation_matrix=nil
raise(NotImplementedError, "only implemented for dense storage")\
unless self.stype == :dense
raise(NotImplementedError, "matrix is not 2-dimensional")\
unless self.dimensions == 2
t = self.clone
pivot = create_dummy_nmatrix
twoDMat = LUDecomposition.new(self.twoDMat).getP
pivot.s = ArrayRealVector.new(ArrayGenerator.getArrayDouble\
(twoDMat.getData, @shape[0], @shape[1]))
return [t,pivot]
end
QRFactorization
def factorize_qr
raise(NotImplementedError, "only implemented for dense storage")\
unless self.stype == :dense
raise(ShapeError, "Input must be a 2-dimensional matrix to have\
a QR decomposition") unless self.dim == 2
qrdecomp = QRDecomposition.new(self.twoDMat)
qmat = create_dummy_nmatrix
qtwoDMat = qrdecomp.getQ
qmat.s = ArrayRealVector.new(ArrayGenerator.\
getArrayDouble(qtwoDMat.getData, @shape[0], @shape[1]))
rmat = create_dummy_nmatrix
rtwoDMat = qrdecomp.getR
rmat.s = ArrayRealVector.new(ArrayGenerator.\
getArrayDouble(rtwoDMat.getData, @shape[0], @shape[1]))
return [qmat,rmat]
end
NMatrix#solve
The solve method currently uses LUDecomposition and Cholesky Decomposition for solving the equations.
def solve(b, opts = {})
raise(ShapeError, "Must be called on square matrix")\
unless self.dim == 2 && self.shape[0] == self.shape[1]
raise(ShapeError, "number of rows of b must equal number\
of cols of self") if self.shape[1] != b.shape[0]
raise(ArgumentError, "only works with dense matrices") if self.stype != :dense
raise(ArgumentError, "only works for non-integer, non-object dtypes")\
if integer_dtype? or object_dtype? or b.integer_dtype? or b.object_dtype?
opts = { form: :general }.merge(opts)
x = b.clone
n = self.shape[0]
nrhs = b.shape[1]
nmatrix = create_dummy_nmatrix
case opts[:form]
when :general, :upper_tri, :upper_triangular, :lower_tri, :lower_triangular
#LU solver
solver = LUDecomposition.new(self.twoDMat).getSolver
nmatrix.s = solver.solve(b.s)
return nmatrix
when :pos_def, :positive_definite
solver = Choleskyecomposition.new(self.twoDMat).getSolver
nmatrix.s = solver.solve(b.s)
return nmatrix
else
raise(ArgumentError, "#{opts[:form]} is not a valid form option")
end
end
NMatrix#matrix_solve
Given we need to solved a system of linear equations
AX = B where A is an m×n matrix, B and X are n×p matrices, we needed to solve this equation by iterating through B.
NMatrix-MRI implements this functionality using NMatrix::BLAS::cblas_trsm operator. However, for NMatrix-JRuby, we implemented NMatrix#matrix_solve.
def matrix_solve rhs
if rhs.shape[1] > 1
nmatrix = NMatrix.new :copy
nmatrix.shape = rhs.shape
res = []
#Solve a matrix and store the vectors in a matrix
(0...rhs.shape[1]).each do |i|
res << self.solve(rhs.col(i)).s.toArray.to_a
end
#res is in col major format
result = ArrayGenerator.getArrayColMajorDouble \
res.to_java :double, rhs.shape[0], rhs.shape[1]
nmatrix.s = ArrayRealVector.new result
return nmatrix
else
return self.solve rhs
end
end
Currently, Hessenberg transformation for an NMatix has not been implemented.
Other dtypes
We have tried implementing float dtypes using jblas FloatMatrix. We here used jblas instead of commons math as Commons Math uses Field Elements for Floats and we may have faced issues with Reflection and TypeErasure. However, we had issues with precision.
Code Organisation and Deployment
To minimise conflict with the MRI codebase all the ruby code has been placed in /lib/nmatrix/jruby directory. /lib/nmatrix/nmatrix.rb decides whether to load nmatrix.so or nmatrix_jruby.rb after detecting the Ruby Platform.
The added advantage of this is at run-time the ruby interpreter must not decide which function to call. The impact on performance can be seen when running programs which intensively use NMatrix for linear algebraic computations(e.g. mixed-models).
Performance
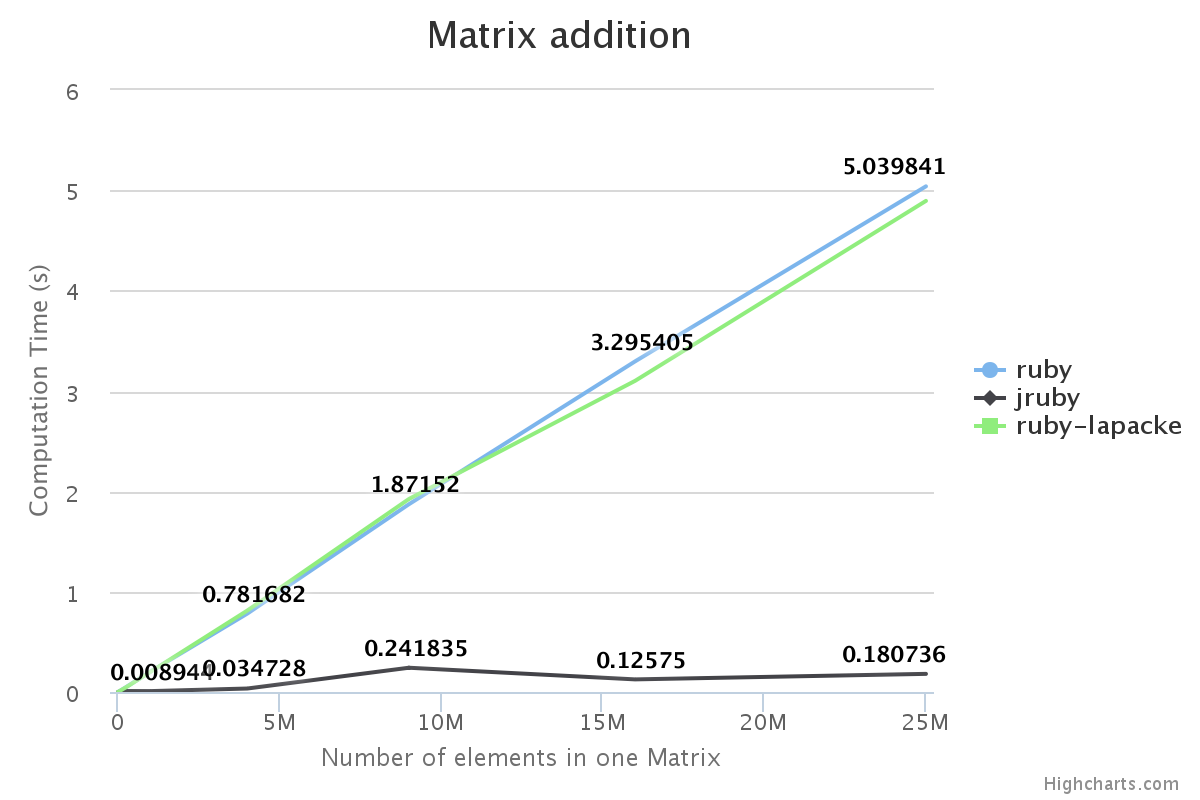
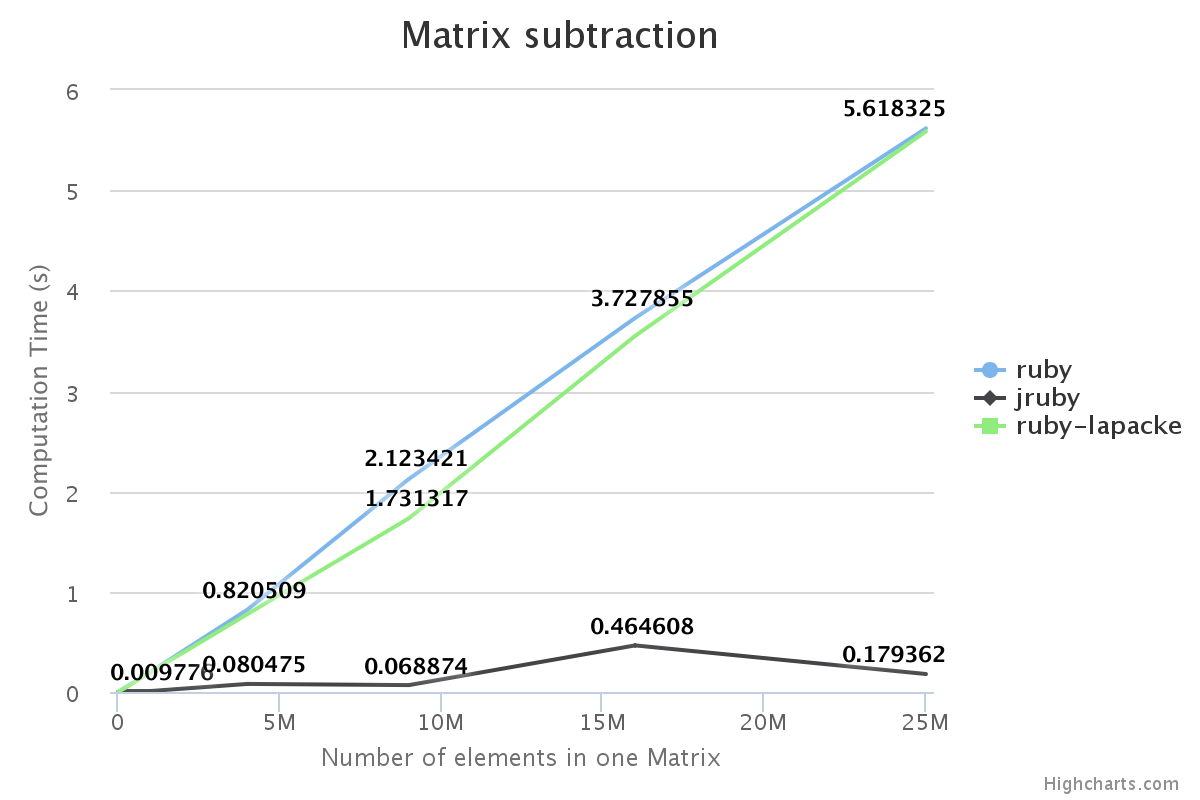
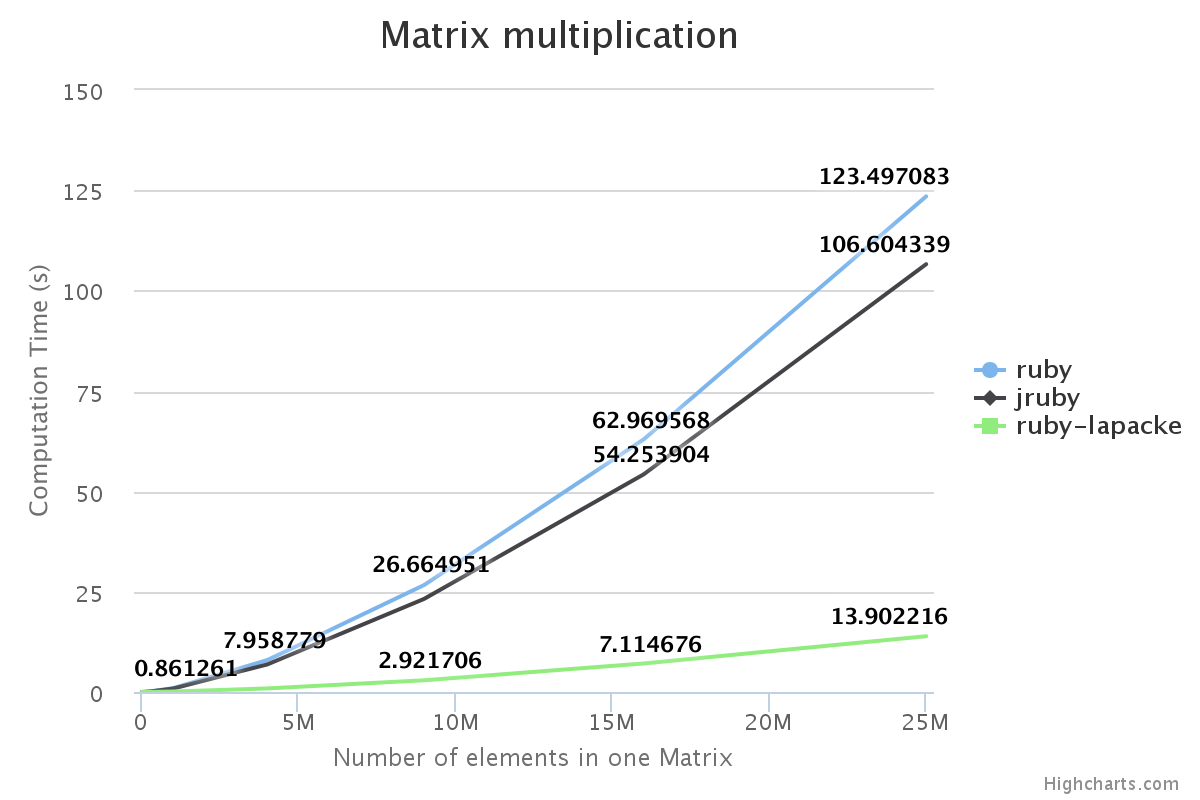
We have benchmarked some of the NMatrix functionalities. The following plots compare the performance between NMatrix-JRuby, NMatrix-MRI and NMatrix-MRI using LAPACKE/ATLAS libraries.
The lower the slope of the curve, the better is the performance.
Note:
- Addition and subtraction are not supported by LAPACKE/ATLAS.
- NMatrix - MRI relies on LAPACKE/ATLAS for calculating determinants and LU Decomposition(lud).
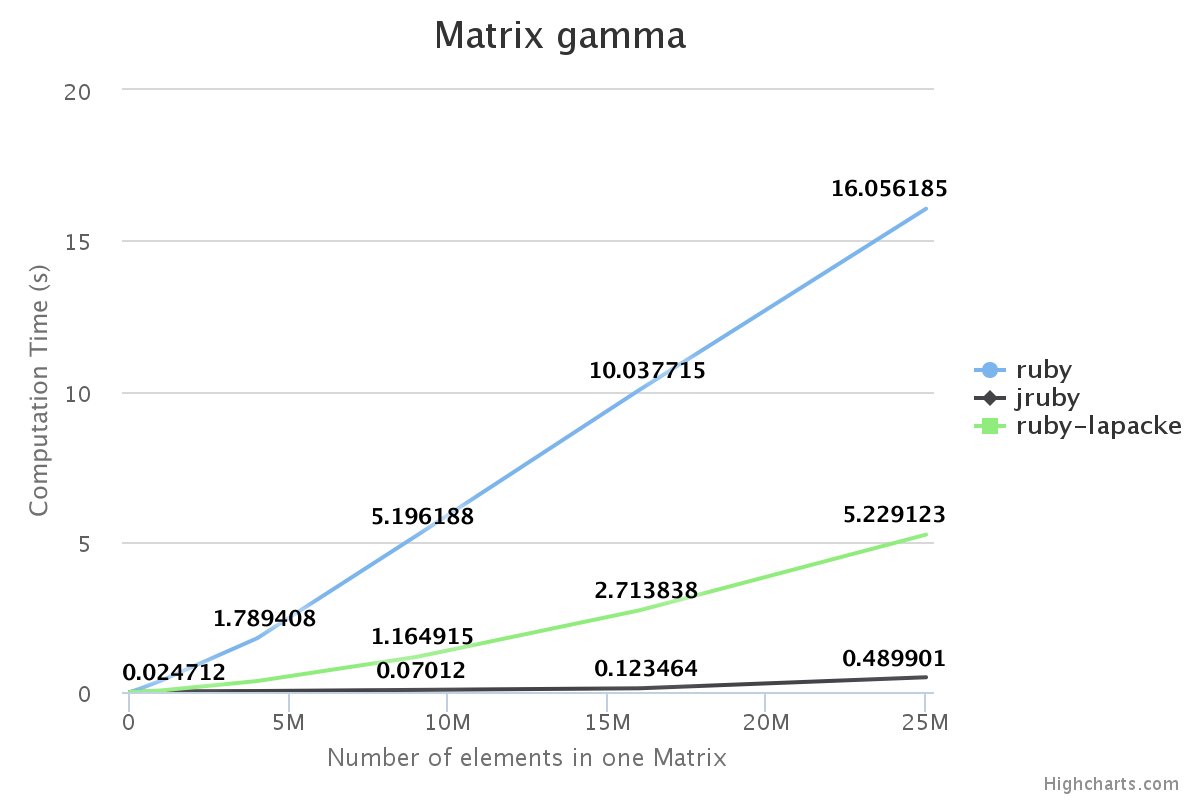
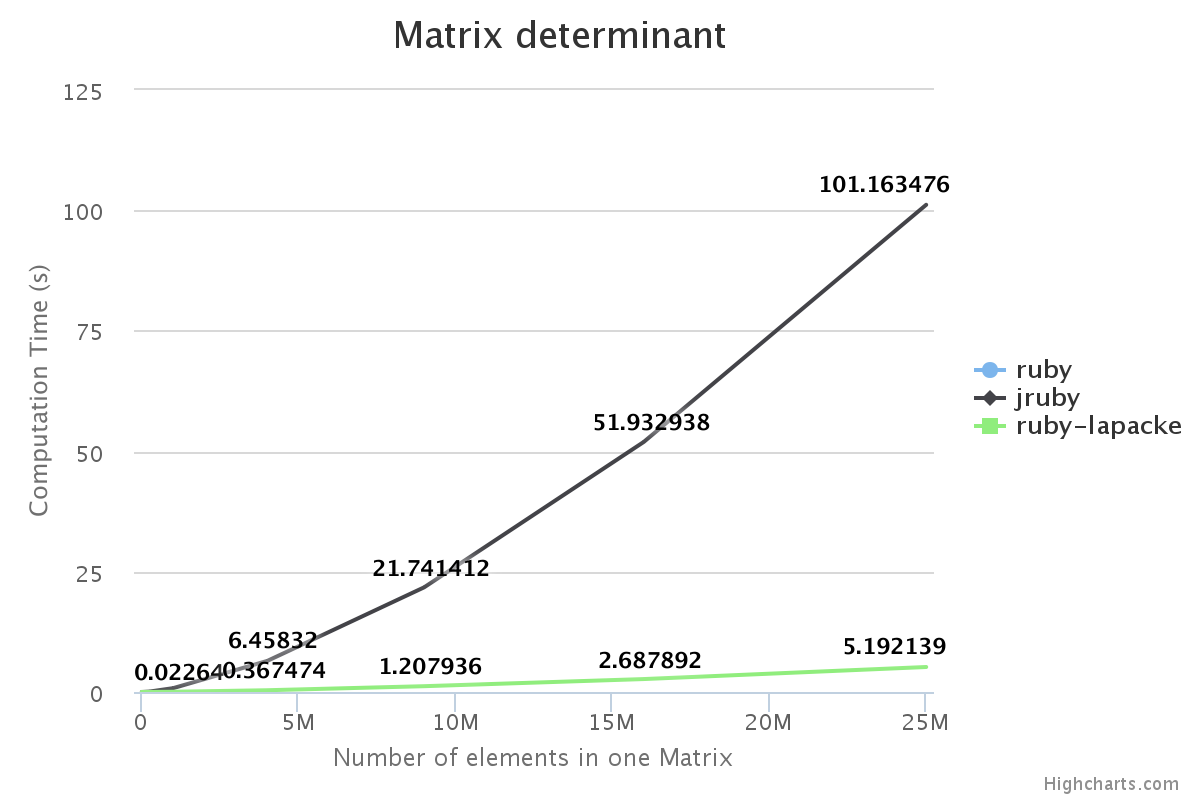
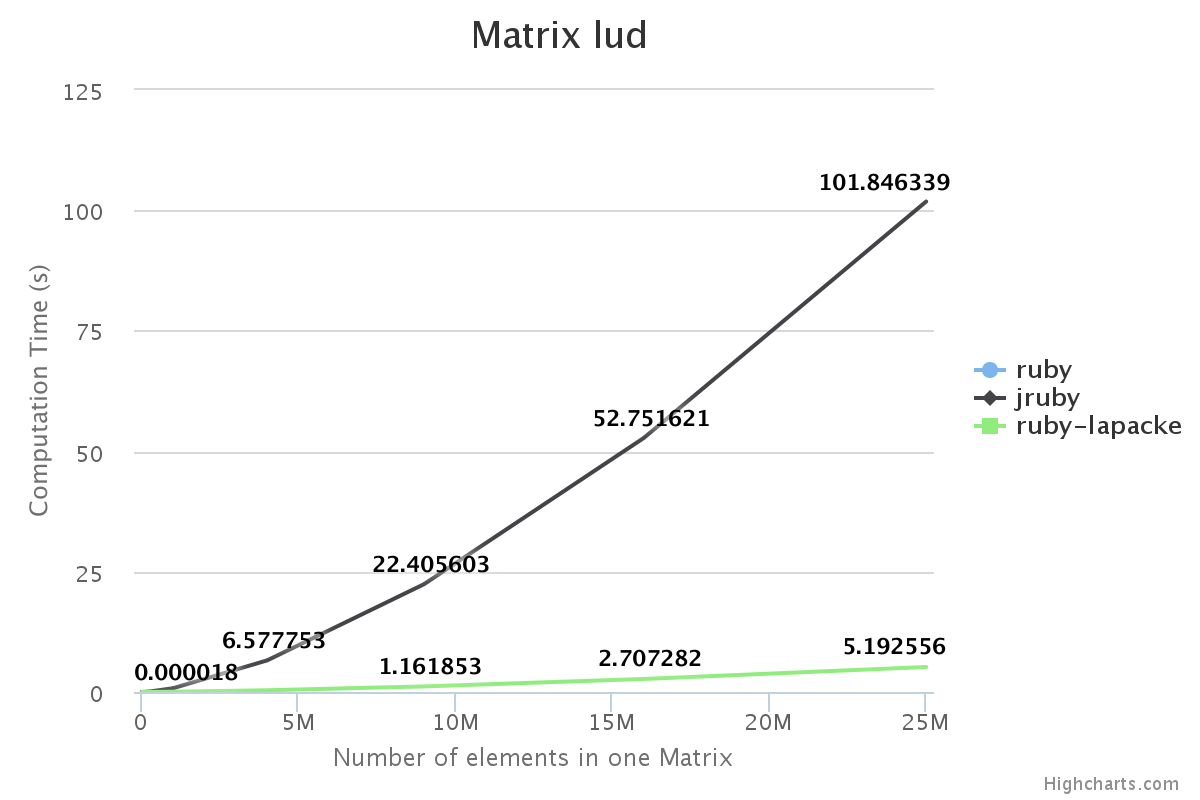
Result:
-
For, two dimensional matrices, NMatrix-JRuby is currently slower than NMatrix-MRI for matrix multiplication, and matrix decomposition functionalities(calculating determinant and factorizing a matrix). NMatrix-JRuby is faster than NMatrix-MRI for other functionalities of a two dimensional matrix, like addition, subtraction, trigonometic operations, etc.
-
NMatrix-JRuby is a clear winner when we are working with matrices of arbitrary dimension.
Test Report
| Spec file | Total Tests | Success | Failure | Pending |
|---|---|---|---|---|
| 00_nmatrix_spec | 188 | 139 | 43 | 6 |
| 01_enum_spec | 17 | 8 | 09 | 0 |
| 02_slice_spec | 144 | 116 | 28 | 0 |
| 03_nmatrix_monkeys_spec | 12 | 11 | 01 | 0 |
| elementwise_spec | 38 | 21 | 17 | 0 |
| homogeneous_spec.rb | 07 | 06 | 01 | 0 |
| math_spec | 737 | 541 | 196 | 0 |
| shortcuts_spec | 81 | 57 | 24 | 0 |
| stat_spec | 72 | 40 | 32 | 0 |
| slice_set_spec | 6 | 2 | 04 | 0 |
Why some tests fail?
- Complex dtype has not been implemented.
- Sparse matrices (list and yale) have not been implemented.
- Decomposition methods that are specific to LAPACK and ATLAS have not been implemented.
- Integer dtype not properly assigned to Floor, Ceil and Round.
Conclusion
The main goal of this project was to bring Scientific Computation to JRuby, to gain from the performance JRuby offers.
By the end of the GSoC, we have been able to successfully create a linear algebra library, NMatrix for JRuby users, which they can easily run on their machines unless they want to use Complex numbers.
We have mixed-models gem simultaneously ported to JRuby. Even here, we are very close to MRI if performance is considered.
Future work
In the coming months we would be implementing Sparse Matrices, thus making NMatrix a complete package for JRuby users. We would also work on improving performance using parallelization. We also feel that JRuby lacks its own Jupyter notebook. The iruby notebook doesn’t work for JRuby. To create an amazing experience for scientific computation on JRuby, we will be porting iruby to JRuby.
Acknowledgements
I would like to express my sincere gratitude to my mentor Pjotr Prins for the continuous support through the summers, and for his patience, motivation, enthusiasm, and immense knowledge. I could not have imagined having a better advisor and mentor, for this project.
I am very grateful to Google and the Ruby Science Foundation for this golden opportunity.
I am very thankful to Charles Nutter, John Woods, Sameer Deshmukh, Kenta Murata and Alexej Gossmann, who mentored me through the project. It has been a great learning experience.
I thank my fellow GSoC participants Rajith, Lokesh and Gaurav who helped me with certain aspects of my project.